Makes a marked point pattern (spatstat.geom::ppp) for the cells
of the specified phenotype in the
specified tissue categories and field.
make_ppp(csd, export_path, pheno, field_name = NULL, tissue_categories = NULL)Arguments
- csd
Cell seg data, may contain multiple fields
- export_path
Path to a directory containing composite and component image files from inForm
- pheno
Phenotype definition. Either a (possibly named) character vector to be parsed by
parse_phenotypes()or a named list containing a single phenotype definition as used byselect_rows().- field_name
Sample Name or Annotation ID for the field of interest. May be omitted if
csdcontains data for only one field.- tissue_categories
Tissue categories of interest. If supplied, the returned
pppwill contain only cells in these categories and the associated window will be restricted to the extent of these categories.
Value
Returns a marked point pattern (spatstat.geom::ppp object)
with a single mark value.
Examples
# ppp for CD8+ cells in the sample data
suppressPackageStartupMessages(library(spatstat))
pp <- make_ppp(sample_cell_seg_data, sample_cell_seg_folder(),
"CD8+", tissue_categories="Tumor")
#> Warning: 6 points were rejected as lying outside the specified window
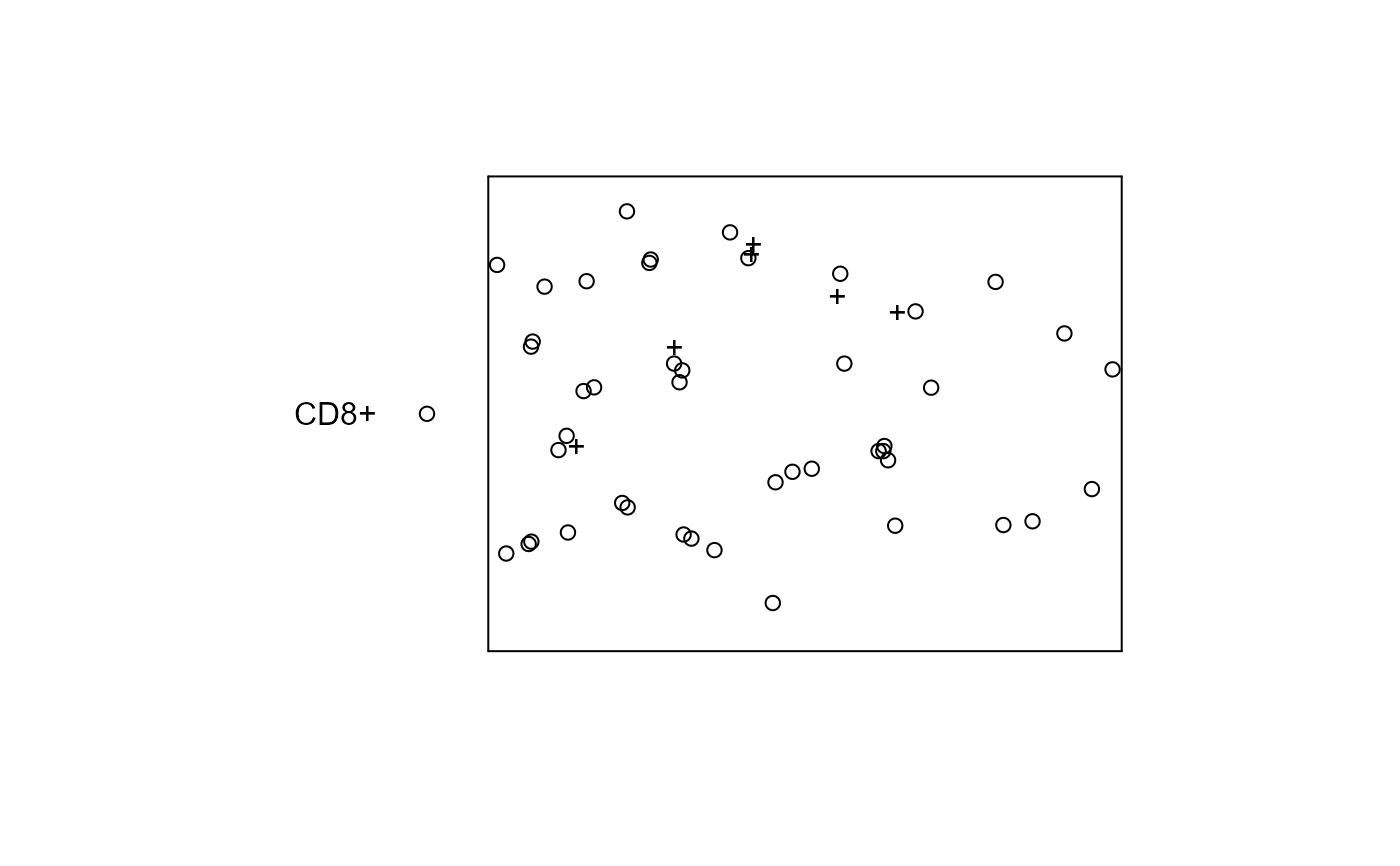
plot(pp, show.window=FALSE, main='')
#> Warning: 6 illegal points also plotted
 # To include multiple phenotypes in a single point pattern,
# create them separately and join them with [spatstat.geom::superimpose()].
pp2 <- make_ppp(sample_cell_seg_data, sample_cell_seg_folder(),
"CK+", tissue_categories="Tumor")
#> Warning: 13 points were rejected as lying outside the specified window
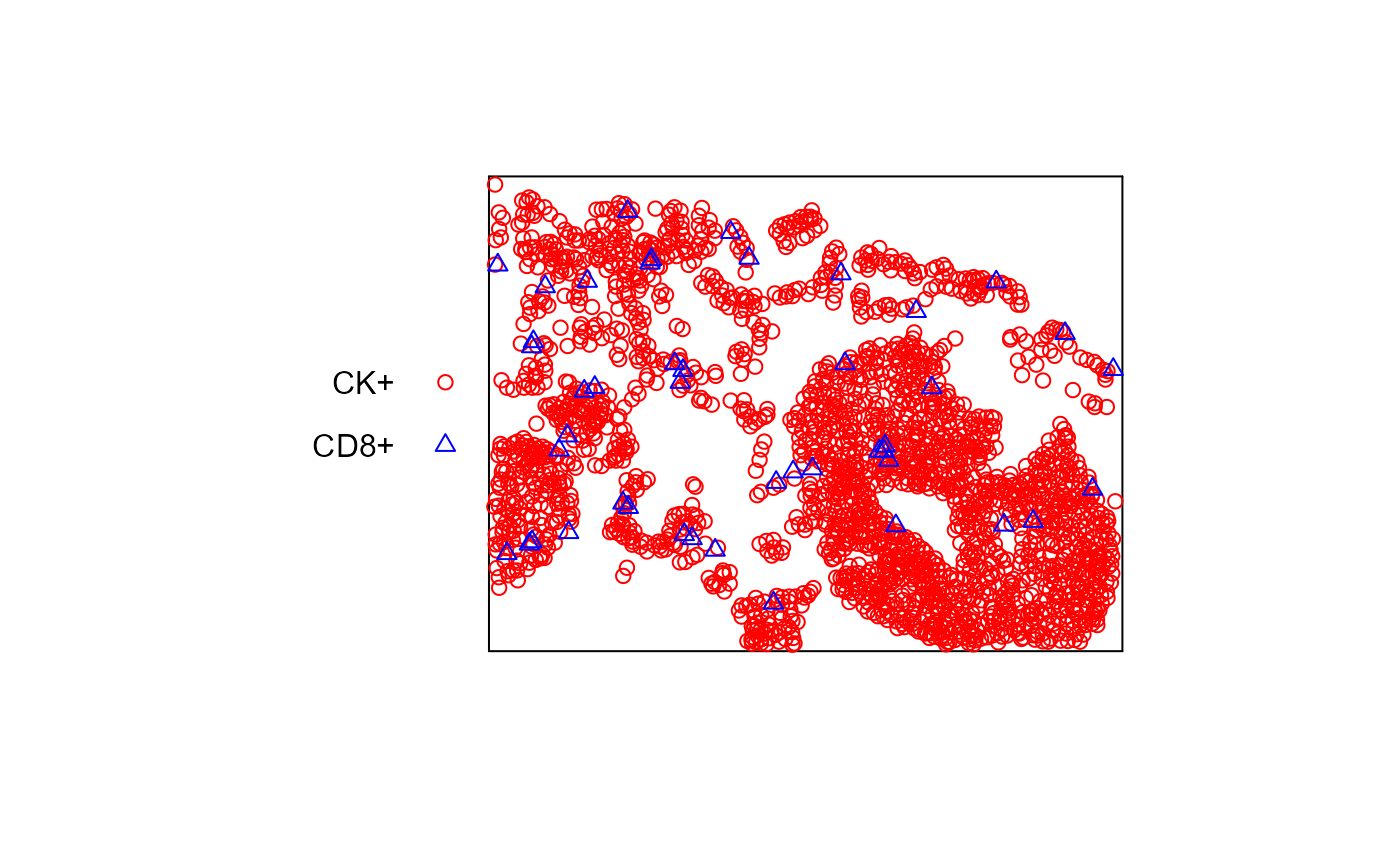
plot(superimpose(pp2, pp), cols=c('red', 'blue'), show.window=FALSE, main='')
# To include multiple phenotypes in a single point pattern,
# create them separately and join them with [spatstat.geom::superimpose()].
pp2 <- make_ppp(sample_cell_seg_data, sample_cell_seg_folder(),
"CK+", tissue_categories="Tumor")
#> Warning: 13 points were rejected as lying outside the specified window
plot(superimpose(pp2, pp), cols=c('red', 'blue'), show.window=FALSE, main='')